Nowadays, Spark surely is one of the most prevalent technologies in the fields of data science and big data. Luckily, even though it is developed in Scala and runs in the Java Virtual Machine (JVM), it comes with Python bindings also known as PySpark, whose API was heavily influenced by Pandas. With respect to functionality, modern PySpark has about the same capabilities as Pandas when it comes to typical ETL and data wrangling, e.g. groupby, aggregations and so on. As a general rule of thumb, one should consider an alternative to Pandas whenever the data set has more than 10,000,000 rows which, depending on the number of columns and data types, translates to about 5-10 GB of memory usage. At that point PySpark might be an option for you that does the job, but of course there are others like for instance Dask which won’t be addressed in this post.
If you are new to Spark, one important thing to note is that Spark has two remarkable features besides its programmatic data wrangling capabilities. One is that Spark comes with SQL as an alternative way of defining queries and the other is Spark MLlib for machine learning. Both topics are beyond the scope of this post but should be taken into account if you are considering PySpark as an alternative to Pandas and scikit-learn for larger data sets.
But enough praise for PySpark, there are still some ugly sides as well as rough edges to it and we want to address some of them here, of course, in a constructive way. First of all, due to its relatively young age, PySpark lacks some features that Pandas provides, for example in areas such as reshaping/pivoting or time series. Also, it is not as straightforward to use advanced mathematical functions from SciPy within PySpark. That’s why sooner or later, you might walk into a scenario where you want to apply some Pandas or SciPy operations to your data frame in PySpark. Unfortunately, there is no built-in mechanism for using Pandas transformations in PySpark. In fact, this requires a lot of boilerplate code with many error-prone details to consider. Therefore we make a wish to the coding fairy, cross two fingers that someone else already solved this and start googling… and here we are ;-)
The remainder of this blog post walks you through the process of writing efficient Pandas UDAFs in PySpark. In fact, we end up abstracting all the necessary boilerplate code into a single Python decorator, which allows us to conveniently specify our PySpark Pandas function. To give more insights into performance considerations, this post also contains a little journey into the internals of PySpark.
UDAFs with RDDs
To start with a recap, an aggregation function is a function that operates on a set of rows and produces a result, for example a sum() or count() function.
A User-Defined Aggregation Function (UDAF) is typically used for more complex aggregations that are not natively shipped with your analysis tool in question.
In our case, this means we provide some Python code that takes a set of rows and produces an aggregate result.
At the time of writing - with PySpark 2.2 as latest version - there is no “official” way of defining an arbitrary UDAF function.
Also, the tracking Jira issue SPARK-10915 does not indicate that this changes in near future.
Depending on your use-case, this might even be a reason to completely discard PySpark as a viable solution.
However, as you might have guessed from the title of this article, there are workarounds to the rescue.
This is where the RDD API comes in.
As a reminder, a Resilient Distributed Dataset (RDD) is the low-level data structure of Spark and a Spark DataFrame is built on top of it. As we are mostly dealing with DataFrames in PySpark, we can get access to the underlying RDD with the help of the rdd attribute and convert it back with toDF().
This RDD API allows us to specify arbitrary Python functions that get executed on the data.
To give an example, let’s say we have a DataFrame df of one billion rows with a boolean is_sold column and we want to filter for rows with sold products. One could accomplish this with the code
df.rdd.filter(lambda x: x.is_sold == True).toDF()
Although not explicitly declared as such, this lambda function is essentially a user-defined function (UDF).
For this exact use case, we could also use the more high-level DataFrame filter() method, producing the same result:
df.filter(df.is_sold == True)
Before we now go into the details on how to implement UDAFs using the RDD API, there is something important to keep in mind which might sound counterintuitive to the title of this post: in PySpark you should avoid all kind of Python UDFs - like RDD functions or data frame UDFs - as much as possible! Whenever there is a built-in DataFrame method available, this will be much faster than its RDD counterpart. To get a better understanding of the substantial performance difference, we will now take a little detour and investigate what happens behind the scenes in those two filter examples.
PySpark internals
PySpark is actually a wrapper around the Spark core written in Scala.
When you start your SparkSession in Python, in the background PySpark uses Py4J to launch a JVM and create a Java SparkContext.
All PySpark operations, for example our df.filter() method call, behind the scenes get translated into corresponding calls on the respective Spark DataFrame object within the JVM SparkContext. This is in general extremely fast and the overhead can be neglected as long as you don’t call the function millions of times.
So in our df.filter() example, the DataFrame operation and the filter condition will be send to the Java SparkContext, where it gets compiled into an overall optimized query plan.
Once the query is executed, the filter condition is evaluated on the distributed DataFrame within Java, without any callback to Python!
In case our workflow loads the DataFrame from Hive and saves the resulting DataFrame as Hive table, throughout the entire query execution all data operations are performed in a distributed fashion within Java Spark workers, which allows Spark to be very fast for queries on large data sets.
Okay, so why is the RDD filter() method then so much slower?
The reason is that the lambda function cannot be directly applied to the DataFrame residing in JVM memory.
What actually happens internally is that Spark spins up Python workers next to the Spark executors on the cluster nodes.
At execution time, the Spark workers send our lambda function to those Python workers.
Next, the Spark workers start serializing their RDD partitions and pipe them to the Python workers via sockets, where our lambda function gets evaluated on each row.
For the resulting rows, the whole serialization/deserialization procedure happens again in the opposite direction so that
the actual filter() can be applied to the result set.
The entire data flow when using arbitrary Python functions in PySpark is also shown in the following image, which has been taken from the old PySpark Internals wiki:
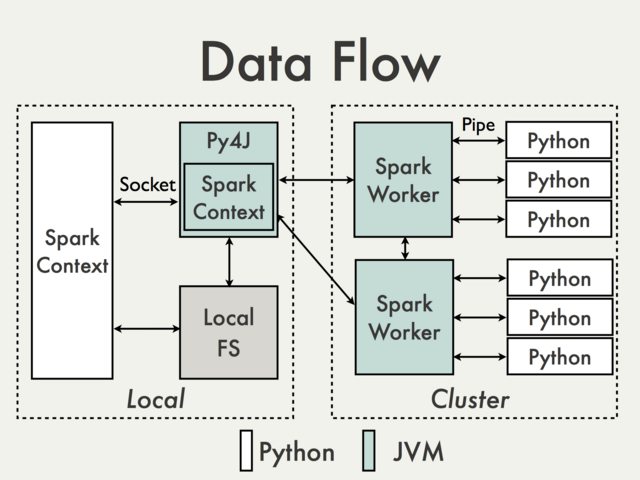
Even if all of this sounded awkwardly technical to you, you get the point that executing Python functions in a distributed Java system is very expensive in terms of execution time due to excessive copying of data back and forth.
To give a short summary to this low-level excursion: as long as we avoid all kind of Python UDFs, a PySpark program will be approximately as fast as Spark program based on Scala. If we cannot avoid UDFs, we should at least try to make them as efficient as possible, which is what we show in the remaining post. Before we move on though, one side note should be kept in mind. The general problem of accessing data frames from different programming languages in the realm of data analytics is currently addressed by the creator of Pandas Wes McKinney. He is also the initiator of the Apache Arrow project which tries to standardize the way columnar data is stored in memory so that everyone using Arrow won’t need to do the cumbersome object translation by serialization and deserialization anymore. Hopefully with version 2.3, as shown in the issues SPARK-13534 and SPARK-21190, Spark will make use of Arrow, which should drastically speed up our Python UDFs. Still, even in that case we should always prefer built-in Spark functions whenever possible.
PySpark UDAFs with Pandas
As mentioned before our detour into the internals of PySpark, for defining an arbitrary UDAF function we need an operation that allows us to operate on multiple rows and produce one or multiple resulting rows.
This functionality is provided by the RDD method mapPartitions, where we can apply an arbitrary Python function my_func to a DataFrame df partition with:
df.rdd.mapPartitions(my_func).toDF()
If you want to further read up on RDDs and partitions, you can checkout the chapter Partitions and Partitioning of the excellent Mastering Apache Spark 2 book by Jacek Laskowski.
In most cases we would want to control the number of partitions, like 100, or even group by a column, let’s say country, in which case we would write:
df.repartition(100).rdd.mapPartitions(my_func).toDF()
or
df.repartition('country').rdd.mapPartitions(my_func).toDF()
Having solved one problem, as it is quite often in life, we have introduced another problem. As we are working now with the low-level RDD interface, our function my_func will be passed an iterator of PySpark Row objects and needs to return them as well. A Row object itself is only a container for the column values in one row, as you might have guessed. When we return such a Row, the data types of these values therein must be interpretable by Spark in order to translate them back to Scala. This is a lot of low-level stuff to deal with since in most cases we would love to implement our UDF/UDAF with the help of Pandas, keeping in mind that one partition should hold less than 10 million rows.
So first we need to define a nice function that will convert a Row iterator into a Pandas DataFrame:
import logging
import pandas as pd
_logger = logging.getLogger(__name__)
def rows_to_pandas(rows):
"""Converts a Spark Row iterator of a partition to a Pandas DataFrame assuming YARN
Args:
rows: iterator over PySpark Row objects
Returns:
Pandas DataFrame
"""
first_row, rows = peek(rows)
if not first_row:
_logger.warning("Spark DataFrame is empty! Returning empty Pandas DataFrame!")
return pd.DataFrame()
first_row_info = ["{} ({}): {}".format(k, rtype(first_row[k]), first_row[k])
for k in first_row.__fields__]
_logger.debug("First partition row: {}".format(first_row_info))
df = pd.DataFrame.from_records(rows, columns=first_row.__fields__)
_logger.debug("Converted partition to DataFrame of shape {} with types:\n{}".format(df.shape, df.dtypes))
return df
This function actually does only one thing which is calling df = pd.DataFrame.from_records(rows, columns=first_row.__fields__) in order to generate a DataFrame. The rest of the code makes sure that the iterator is not empty and for debugging reasons we also peek into the first row and print the value as well as the datatype of each column. This has proven in practice to be extremely helpful in case something goes wrong and one needs to debug what’s going on in the UDF/UDAF. The functions peek and rtype are defined as follows:
from itertools import chain
def peek(iterable):
"""Peek into the first element and return the whole iterator again
Args:
iterable: iterable object like list or iterator
Returns:
tuple of first element and original iterable
"""
iterable = iter(iterable)
try:
first_elem = next(iterable)
except StopIteration:
return None, iterable
iterable = chain([first_elem], iterable)
return first_elem, iterable
def rtype(var):
"""Heuristic representation for nested types/containers
Args:
var: some (nested) variable
Returns:
str: string representation of nested datatype (NA=Not Available)
"""
def etype(x):
return type(x).__name__
if isinstance(var, list):
elem_type = etype(var[0]) if var else "NA"
return "List[{}]".format(elem_type)
elif isinstance(var, dict):
keys = list(var.keys())
if keys:
key = keys[0]
key_type, val_type = etype(key), etype(var[key])
else:
key_type, val_type = "NA", "NA"
return "Dict[{}, {}]".format(key_type, val_type)
elif isinstance(var, tuple):
elem_types = ', '.join(etype(elem) for elem in var)
return "Tuple[{}]".format(elem_types)
else:
return etype(var)
The next part is to actually convert the result of our UDF/UDAF back to an iterator of Row objects. Since our result will most likely be a Pandas DataFrame or Series, we define the following:
import numpy as np
from pyspark.sql.types import Row
def convert_dtypes(rows):
"""Converts some Pandas data types to pure Python data types
Args:
rows (array): numpy recarray holding all rows
Returns:
Iterator over lists of row values
"""
dtype_map = {pd.Timestamp: lambda x: x.to_pydatetime(),
np.datetime64: lambda x: pd.Timestamp(x).to_pydatetime(),
np.bool_: lambda x: bool(x),
np.int8: lambda x: int(x),
np.int16: lambda x: int(x),
np.int32: lambda x: int(x),
np.int64: lambda x: int(x),
np.float16: lambda x: float(x),
np.float32: lambda x: float(x),
np.float64: lambda x: float(x),
np.float128: lambda x: float(x)}
for row in rows:
yield [dtype_map.get(type(elem), lambda x: x)(elem) for elem in row]
def pandas_to_rows(df):
"""Converts Pandas DataFrame to iterator of Row objects
Args:
df: Pandas DataFrame
Returns:
Iterator over PySpark Row objects
"""
if df is None:
_logger.debug("Returning nothing")
return iter([])
if type(df) is pd.Series:
df = df.to_frame().T
if df.empty:
_logger.warning("Pandas DataFrame is empty! Returning nothing!")
return iter([])
_logger.debug("Convert DataFrame of shape {} to partition with types:\n{}".format(df.shape, df.dtypes))
records = df.to_records(index=False)
records = convert_dtypes(records)
first_row, records = peek(records)
first_row_info = ["{} ({}): {}".format(k, rtype(v), v) for k, v in zip(df.columns, first_row)]
_logger.debug("First record row: {}".format(first_row_info))
row = Row(*df.columns)
return (row(*elems) for elems in records)
This looks a bit more complicated but essentially we convert a Pandas Series to a DataFrame if necessary and handle the edge cases of an empty DataFrame or None as return value. We then convert the DataFrame to records, convert some NumPy data types to the Python equivalent and create an iterator over Row objects from the converted records.
With these functions at hand we can define a Python decorator that will allow us to automatically call the functions rows_to_pandas and pandas_to_rows at the right time:
from functools import wraps
class pandas_udaf(object):
"""Decorator for PySpark UDAFs using Pandas
Args:
loglevel (int): minimum loglevel for emitting messages
"""
def __init__(self, loglevel=logging.INFO):
self.loglevel = loglevel
def __call__(self, func):
@wraps(func)
def wrapper(*args):
# use *args to allow decorating methods (incl. self arg)
args = list(args)
setup_logger(loglevel=self.loglevel)
args[-1] = rows_to_pandas(args[-1])
df = func(*args)
return pandas_to_rows(df)
return wrapper
The code is pretty much self-explanatory if you have ever written a Python decorator; otherwise, you should read about it since it takes some time to wrap your head around it. Basically, we set up a default logger, create a Pandas DataFrame from the Row iterator, pass it to our UDF/UDAF and convert its return value back to a Row iterator. The only additional thing that might still raise questions is the usage of args[-1]. This is due to the fact that func might also be a method of an object. In this case, the first argument would be self but the last argument is in either cases the actual argument that mapPartitions will pass to us. The code of setup_logger depends on your Spark installation. In case you are using Spark on Apache YARN, it might look like this:
import os
import sys
def setup_logger(loglevel=logging.INFO, logfile="pyspark.log"):
"""Setup basic logging for logging on the executor
Args:
loglevel (int): minimum loglevel for emitting messages
logfile (str): name of the logfile
"""
logformat = "%(asctime)s %(levelname)s %(module)s.%(funcName)s: %(message)s"
datefmt = "%y/%m/%d %H:%M:%S"
try:
logfile = os.path.join(os.environ['LOG_DIRS'].split(',')[0], logfile)
except (KeyError, IndexError):
logging.basicConfig(level=loglevel,
stream=sys.stdout,
format=logformat,
datefmt=datefmt)
logger = logging.getLogger(__name__)
logger.error("LOG_DIRS is not in environment variables or empty, using STDOUT instead.")
logging.basicConfig(level=loglevel,
filename=logfile,
format=logformat,
datefmt=datefmt)
Now having all parts in place let’s assume the code above resides in the python module pyspark_udaf.py. A future post will cover the topic of deploying dependencies in a systematic way for production requirements. For now we just presume that pyspark_udaf.py as well as all its dependencies like Pandas, NumPy, etc. are accessible by the Spark driver as well as the executors. This allows us to then easily define an example UDAF my_func that collects some basic statistics for each country as:
import pyspark_udaf
import logging
@pyspark_udaf.pandas_udaf(loglevel=logging.DEBUG)
def my_func(df):
if df.empty:
return
df = df.groupby('country').apply(lambda x: x.drop('country', axis=1).describe())
return df.reset_index()
It is of course not really useful in practice to return some statistics with the help of a UDAF that could also be retrieved with basic PySpark functionality but this is just an example. We now generate a dummy data DataFrame and apply the function to each partition as above with:
# make pyspark_udaf.py available to the executors
spark.sparkContext.addFile('./pyspark_udaf.py')
df = spark.createDataFrame(
data = [('DEU', 2, 1.0), ('DEU', 3, 8.0), ('FRA', 2, 6.0),
('FRA', 0, 8.0), ('DEU', 3, 8.0), ('FRA', 1, 3.0)],
schema = ['country', 'feature1', 'feature2'])
stats_df = df.repartition('country').rdd.mapPartitions(my_func).toDF()
print(stats_df.toPandas())
The code above can be easily tested with the help of a Jupyter notebook with PySpark where the SparkSession spark is predefined.
Summary
Overall, this proposed method allows the definition of an UDF as well as an UDAF since it is up to the function my_func if it returns (1) a DataFrame having as many rows as the input DataFrame (think Pandas transform), (2) a DataFrame of only a single row or (3) optionally a Series (think Pandas aggregate) or a DataFrame with an arbitrary number of rows (think Pandas apply) with even varying columns.
Therefore, this approach should be applicable to a variety of use cases where the built-in PySpark functionality is not sufficient.
To wrap it up, this blog post gives you a template on how to write PySpark UD(A)Fs while abstracting all the boilerplate in a dedicated module. We also went down the rabbit hole to explore the technical difficulties the Spark developers face in providing Python bindings to a distributed JVM-based system. In this respect we are really looking forward to closer integration of Apache Arrow and Spark in the upcoming Spark 2.3 and future versions.
This article was coauthored by my inovex colleague Bernhard Schäfer and was also published on the inovex blog.
Comments
comments powered by Disqus